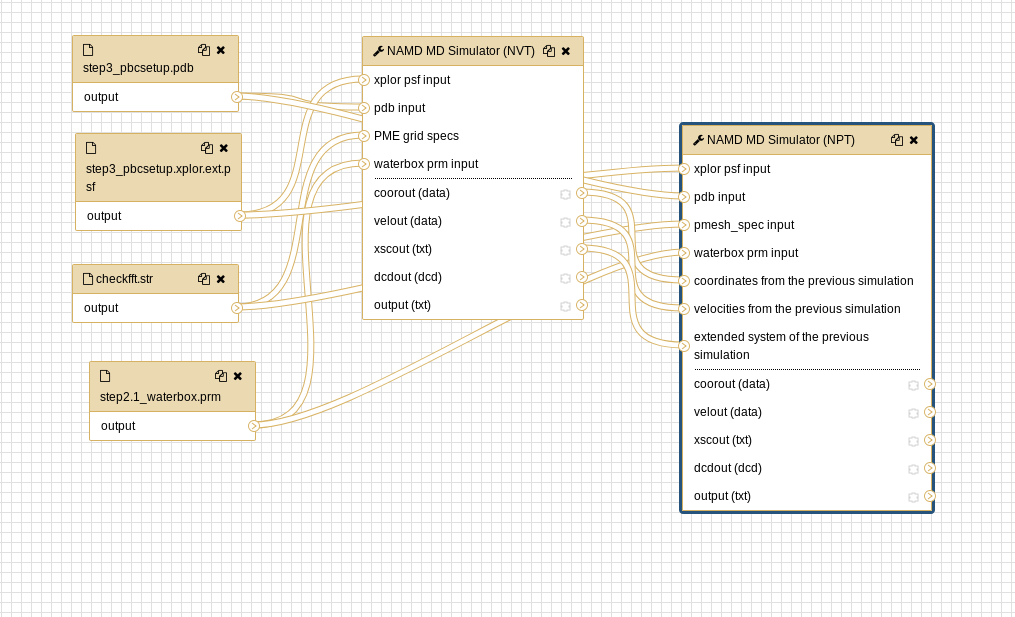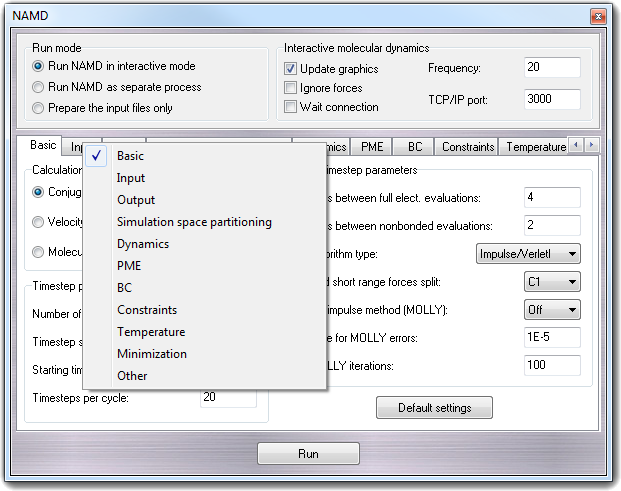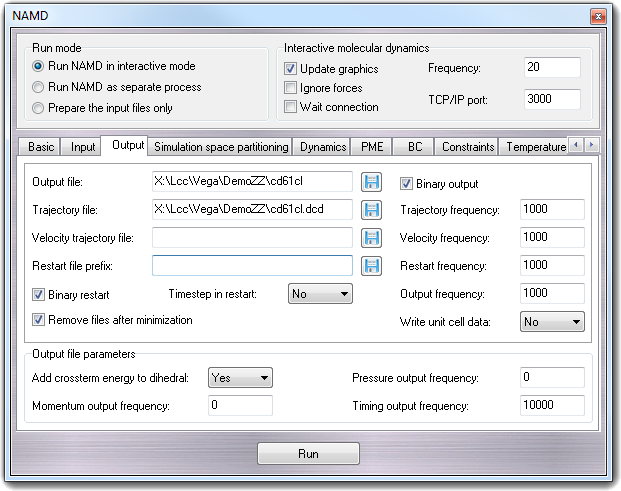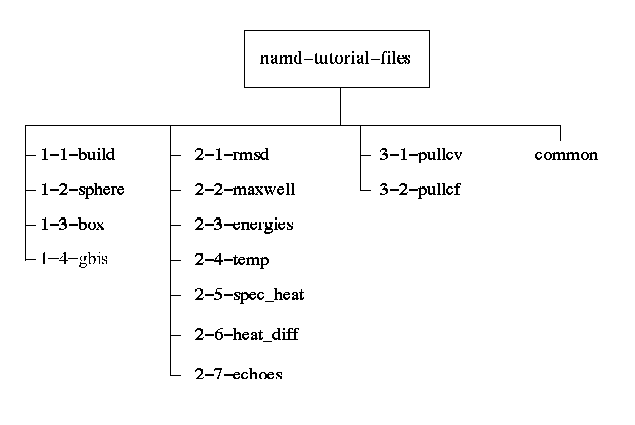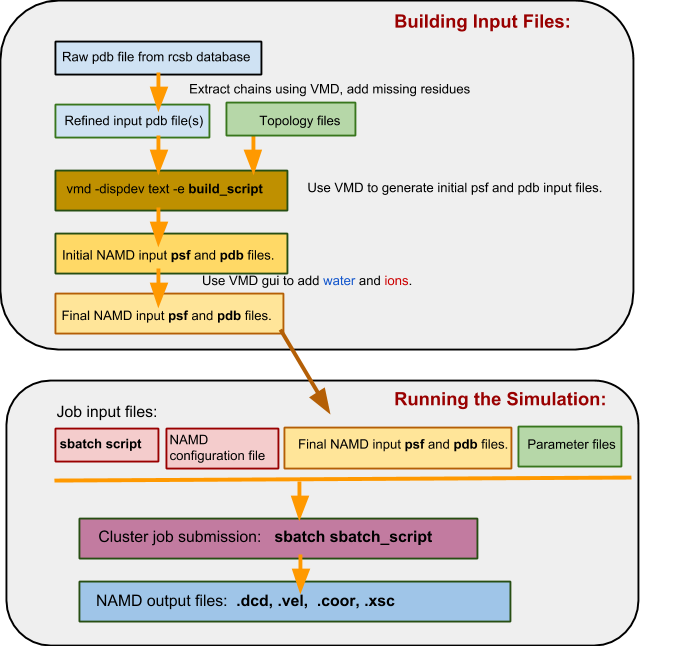
Molecular Dynamics - Building input files, visualising the trajectory - Bioinformatics Documentation

Girinath G. Pillai on Twitter: "Do you want to run Molecular Dynamics via a web browser? Run MD simulation in @GoogleColab using #NAMD GPU Colab Link: https://t.co/Zde7xBVYBj Video demo: https://t.co/8n68d9hnHT For learning

Why do I get an errro when setting up a simulation of my protein-ligand complex by NAMD using its visual interface VMD? | ResearchGate

GitHub - quantaosun/NAMD-MD: Open Source, Prepare a molecular dynamic simulation from scratch without installing local VMD, use LigParGen for topology, it is one of the most reliable MD for a general solution